EvoSimulator 2
EvoSimulator: a simple tool illustrating the power of ‘mutation + selection’
The ‘EvoSimulator’ program makes it possible to experience in a simple way (by means of numerical simulations) the relative importance of basic evolutionary mechanisms (mutation, selection and recombination) in the emergence of complexity.
Download
- Windows version (english | french)
- MacOS version (english | french)
- Linux version (english | french)
How to use the EvoSimulator' ?
The individuals of the simulated species are represented by their ‘genome’ of fixed size and composed of characters. Instead of using a 4-letter alphabet (A, C, G, and T), we use a much richer alphabet (81 possible characters) covering all the letters of the Latin alphabet (upper and lower case letters, with or without accents), the 10 digits and some special characters (including the 'blank'): [aàAbBcçCdDéeèEfFgGhHiIjJkKlLmMnNoOpPqQrRsStTuùUvVwWxXyYzZ 0123456789,.?!'\"()].
The number of different possible sentences (genomes) depends on the size of the target sentence (chosen by the user). By default, the target genome is: This sentence is COMPLEX and cannot emerge by chance.
This sentence is 52-character long. The possible number of genomes of 52 characters using an alphabet of 81 possible characters is equal to 81 ^ 52> 1.7x10 ^ 99 ... that is, a number very much greater than the number of particles in the universe. It would take virtually infinite time for a computer to generate all possible sentences and therefore generate the target sentence with certainty. Chance alone cannot generate complexity.
… But everything changes if we bring in the mechanism of Darwinian selection. The fitness of each individual is measured objectively by the proximity of the sequence of characters to a "target sentence" (of maximum fitness). To do this, EvoSimulator calculates, for each generated genome, a score which represents the proportion of characters identical to the target genome. Note that a character is only identical if it is also in the same position in the sentence. For example, if an individual has a score of 60%, this means that 60% of the positions in its genome are identical to the characters at the same positions in the target genome and that 40% are different. Note that "a", "à" and "A" are different characters!
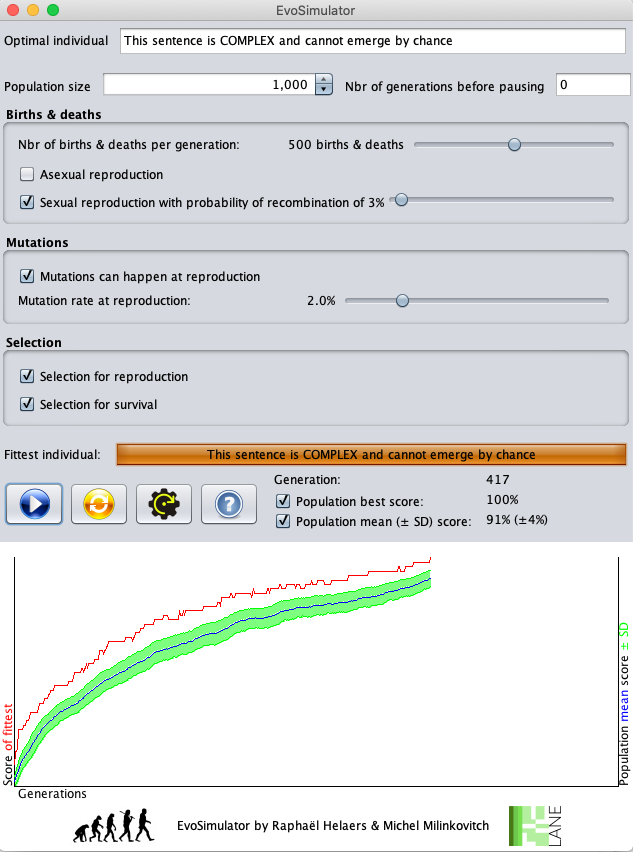
Before starting a simulation, the population size (by default 1000 individuals) must be determined. It will remain fixed over time because, at each new generation, the number of individuals that will die will be the same as the number of individuals that will get born.
The parameters that you can control are the following:
- Panel ‘Births & deaths’
At each generation, a certain number of deaths and births take place (by default 500, the slider "Number of births & deaths per generation" allows you to modify this value). As previously indicated, there will be as many births as there are deaths, so that the population size remains stable. In the same panel, you can control reproduction type: asexual (an individual who reproduces creates a copy of itself) or sexual with recombination (2 individuals are chosen as parents and give birth to a child whose genome will be a mixture of the two parental genomes). - Panel ‘Mutations’
During reproduction, the genome of the parent(s) is (are) copied character by character to form the new individual (i.e., the child). If you check the ‘mutations’ box, there is a probability (by default 2%, the "mutation rate" slider allows this value to be modified) for each copied character to write a random character instead. If you uncheck the box, no mutation will be generated, and the characters will always be copied without error. - Panel ‘Selection’
Selection can be applied to the choice of individuals who reproduce and / or to the choice of individuals who die (depending on the box(es) checked). If selection is applied during reproduction, each individual has a probability proportional to its score to be chosen to reproduce, otherwise it is chosen at random. If selection is applied during survival, each individual has a probability inversely proportional to its score to be chosen to die, otherwise it is chosen at random.
The controls on the currently running simulation are as follows:
- At each generation, the genome (= sentence) of the fittest individual is displayed with its score (the box is filled in proportion to the relative score). The graph saves this best score for the corresponding generation.
- During the simulation, the user can press the start/pause button to resume/pause the run.
- During the simulation, the user can modify any parameter so that it is immediately taken into account (with the exception of the population size, which must be modified in pause mode).
- The second button is used to restart the simulation at the generation zero.
- The third button allows you to reset the default settings.
- The box "Nbr of generations before pausing" next to the population size panel allows automatically pausing the simulation every X generations.
References and links
- The simulation of mutation and selection on a population of sentences has been suggested by Richard Dawkins in his book “The blind watchmaker” and implemented in a program called “the weasel program”.
- Why is the ID thesis incorrect and how this approach is inconsistent with the foundations of the scientific method?
The University Journal (in French)
Flyer (in French) - Understanding Evolution, your one-stop source for information on evolution (University of Berkeley)
- Evolution resources from the National Academies
- PhyloIntelligence, a site summarizing some of the evidence for evolution
- NOVA TV programs on evolution